CPS Meeting 2019
Please find informations on our final project report meeting here:
Conference report:
PDF-File (1,7 mB)
Conference website:
www.cps2019.de
1.1.4. Enzyme-catalyzed ligations
Efficient enzymatic methods provide an alternative to chemical ligation strategies (see 1.1.1) in the production of important starting materials for ligation by exploiting the high chemoselectivity of enzymes in the biosynthesis of peptides.
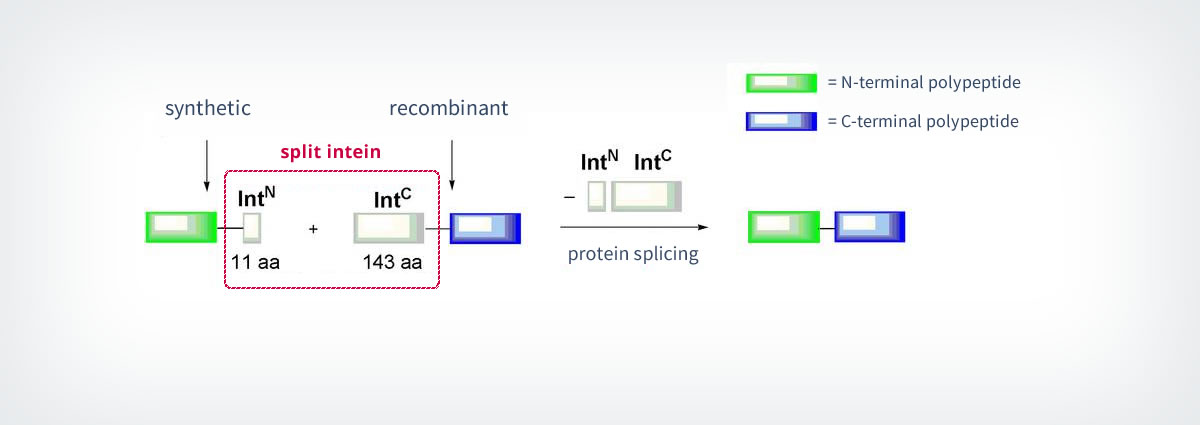
The most commonly known methods use inteins for the so-called “protein trans-splicing”. The splicing process is an important aspect of Expressed Protein Ligation (EPL, see part 1.1.1), because it is used to generate the needed recombinant thioesters for the ligation. Protein splicing is a naturally occurring event during which the auto-processing protein domain (“Intein”) excises itself out of a precursor polypeptide and links the adjacent N- and C-terminal fragments (“Exteins”) by a native peptide bond34. The intein catalyses all the enzymatic reactions needed for this process. Besides the normal intein domains, nature also features inteins that are split into two fragments. Split-inteins are associated into a functional complex and catalyse the splicing between polypeptides (Scheme 5). In some cases, split-inteins can also be generated artificially by splitting regular intein domains into two. The artificially split Ssp-DnaB intein illustrates nicely the usefulness of these constructs: The N-terminal fragment of the split-intein (IntN) contains only 11 amino acids and is consequently not only quick to synthesise, but also easy to modify by elongation with other amino acid residues. The IntN is then linked to the C-terminal fragment (IntC) to yield the final product.35 Protein splicing only needs low concentrations of the reactants. Other advantages are the lack of special functional groups and the possibility of using the reaction in living cells.36
Another important enzyme-based methodology is reverse proteolysis, which is defined by the reversal of the protease-catalysed peptide bond hydrolysis. Here the enzyme, the reaction conditions or the reactants are changed in such a way that the proteases function as protein ligases. In the simplest of all cases, the reaction is carried out in an organic medium, which will push the thermodynamic equilibrium between proteolysis and reverse proteolysis to reverse proteolysis (thermodynamically controlled reverse proteolysis). However, the use of organic medium drastically reduces the enzyme’s activity. During the kinetically controlled reverse proteolysis, active peptide substrates are used to generate an artificial peptide-protease intermediate, which can be reacted off with another peptide or protein. This principle is especially useful for the semi-synthesis or the selective modification of proteins.37 The expressed enzymatic ligation (EEL) combines the principle of reverse proteolysis with EPL and has been successfully used for the synthesis of the pheromone neuropeptide Y from two peptide fragments.38
A further example of enzyme-catalyzed ligation is the activity of transpeptidase sortase A, which catalyses peptide ligations in nature. To be able to use sortase A in a ligation, the N-terminal peptide has to contain an LPxTG motif at the C-terminus. In addition, the C-terminal fragment also has to be equipped with a Gly residue at its N-terminus.39 Sortase A like cleave off the so-called “sorting motif” of the N-terminal peptide during the ligation and will then link it to the Gly residue at the N-terminus of the C-terminal peptide by the formation of a natural peptide bond.
Besides the just mentioned applications of enzyme-catalyzed ligations, they can also be used to modify proteins. Two examples include the selective labelling of proteins with fluorophores and protein immobilization. In addition, enzymes-catalyzed ligations can be combined nicely with orthogonal functionalization strategies40, as demonstrated with the SNAP tag, where a benzylguanine derivative is covalently liked to a protein by the fused O6-alkylguanine-DNA-alkyltransferase (hAGT).41 Another example is the Halo-tag, where chlorohexane is conjugated by haloalkane-dehalogenases.42